This article provides an introductory guide to navigating the FlagEmbedding library, covering sequential processes such as hard negative mining, fine-tuning, and evaluation to develop a customized embedding model. It will also share key insights gained through troubleshooting and experimentation.
1. Why FlagEmbedding?
FlagEmbedding is a toolkit developed by BAAI that supports one-stop retrieval for search and RAG. I specifically used this tool because of multiple reasons:
1.
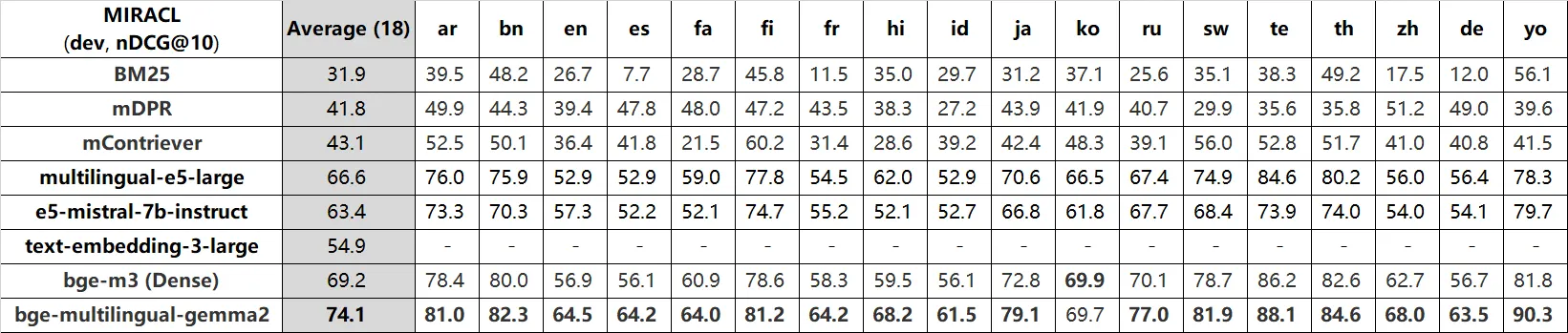
Referring to the benchmark below, bge-m3 displays a high performance in Korean, even comparatively to bge-multilingual-gemma2. FlagEmbedding provides models of the BGE (BAAI General Embeddings) series that achieve top rankings.
2.
As it was inferred, FlagEmbedding supports models that are available for multiple languages. It is especially structured around English and Chinese.
3.
Their research is ongoing and active; they actually have made huge changes in the github very recently. This easily makes troubleshooting accessible.
2. Installation process
Personally, I used FlagEmbedding for the purpose of my research in Hard Negative Mining and the entire procedure was done on Docker as it effectively maintains an isolated environment for the FlagEmbedding library.
1.
Cloning FlagEmbedding and installing required libraries
git clone https://github.com/FlagEmbedding/FlagEmbedding.git
cd FlagEmbedding
pip install -e .[finetune]
Bash
복사
2.
Install pytrec_eval and faiss for evaluation of models
pip install pytrec_eval
pip install https://github.com/kyamagu/faiss-wheels/releases/download/v1.7.3/faiss_gpu-1.7.3-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
Python
복사
Error while creating shared memory segment
•
On installation, if you are using Docker, this is an example configuration for the docker run command that bypasses the shared memory error.
docker run --shm-size=4g -it -d --gpus all --name flagembed -v $(pwd):/workspace <IMAGE_NAME>
Bash
복사
3. Data Preparation with Hard Negative Mining
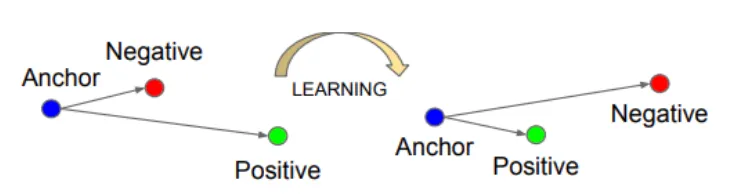
Generally, embedding models are finetuned through contrastive learning. Contrastive learning is a type of self-supervised learning where models learn the distance between distinct data points. Within contrastive learning there are three key components; anchor (query), positive (answer document), and negative (irrelevant or insufficient document). Simply, we want the model to learn to position the positive data points nearby the query and negative data points further away.
Thus, to train our embedding models, we need a dataset in this example structure:
{
"query": "What is the capital city of Korea?",
"pos": ["Seoul is the capital city of Korea"],
"neg": ["Gangnam is the most beautiful city in Korea", "Noodles are the best late-night snack"]
}
JSON
복사
Not all negatives are the same. Looking at the example data above, you can see that one is completely unrelated to the query, while another is quite relevant to it. For instance, if the query is asking about "the capital of Korea," the sentence "Noodles are the best late-night snack" is entirely unrelated to the query. Just as we can intuitively recognize that these two sentences are completely unrelated, embedding models are also likely to perceive this difference and place the query and this sentence far apart when vectorizing them.
On the other hand, the sentence "Gangnam is the most beautiful city in Korea" is somewhat related to the query. Since it shares the common topic of "city," the vector for this sentence is more likely to be positioned closer to the query.
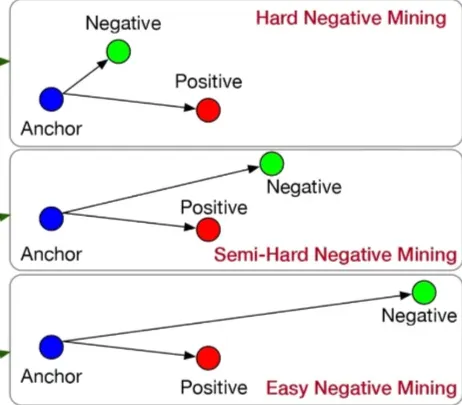
There are levels to the difficulties of the negatives. Some are easily distinguishable and trivial to answering the query, while others are quite relevant and difficult to determine whether it is sufficient to answer the question. We label the latter as hard negatives. Usually, easy negatives are already placed far away from the anchor within the embedding model’s vector space. To maximize performance increase, it is important to have hard negatives that are similar within the base model’s vector space, so that throughout contrastive learning, the model learns to distinguish and emphasize the differences between the two.
Hard Negative Mining (HNM) ensures that the negative documents for training data are difficult and similar to the query; ideally falling just short of the answer. Although there are various implementation of HNM strategies, FlagEmbedding already has a simple dense retrieval HNM script that suffice.
python scripts/hn_mine.py \
--model_name_or_path BAAI/bge-multilingual-gemma2 \
--input_file toy_finetune_data.jsonl \
--output_file toy_finetune_data_minedHN.jsonl \
--range_for_sampling 2-200 \
--negative_number 15 \
--use_gpu_for_searching
Bash
복사
•
There are scripts for splitting data and assigning reranker teacher scores as well.  GitHubFlagEmbedding/scripts at master · FlagOpen/FlagEmbedding
GitHubFlagEmbedding/scripts at master · FlagOpen/FlagEmbedding
•
range_for_sampling : 2-200 means sampling negatives from top 2 to top 200 documents. larger value will reduce the difficulty of negatives.
•
negative_number : the number of negatives can be scaled according to the dataset
•
For my case, HNM scripts were run on QA datasets, where the answers of the entire QA dataset become the corpus where the hard negatives are mined from. This results in a single positive document per query.
If this is successfully run, we’ll have a dataset of queries and its corresponding 15 hard negatives that have been randomly sampled from the top 2-200 most similar documents from the corpus by bge-multilingual-gemma2.
Note that if you desire to evaluate your model on this data, you should split the data at this point.
4. BGE-M3 FineTuning
Note that this command is specific to finetuning bge-m3. For other others, refer to the documentation.
•
train_group_size determines the number of data samples grouped in one training batch. query_max_len and passage_max_len sets the maximum token length for quries and passages. These can be manipulated according to the data and resource availability.
•
knowledge_distillation is set to False because we didn’t assign any teacher scores and won’t be leveraging model distillation.
•
same_dataset_within_batch specifies whether the same dataset should be used within a batch. Setting this to False increases data diversity.
•
We are using self-distillation during unify fine-tuning, starting at step 500. This involves the model using its own predictions as additional training signals to refine its embeddings. In this setup, self-distillation starts at step 500, allowing the model to first learn from the original ground truth before incorporating its own outputs.
CUDA_VISIBLE_DEVICES="1,2,3,4,5,6,7" \
torchrun --nproc_per_node 5 \
-m FlagEmbedding.finetune.embedder.encoder_only.m3 \
--model_name_or_path BAAI/bge-m3 \
--cache_dir ./cache/model \
--train_data toy_finetune_data_minedHN.jsonl \
toy_finetune_data_minedHN2.jsonl \
toy_finetune_data_minedHN3.jsonl \
--cache_path ./cache/data \
--train_group_size 8 \
--query_max_len 128 \
--passage_max_len 400 \
--pad_to_multiple_of 8 \
--knowledge_distillation False \
--same_dataset_within_batch False \
--small_threshold 0 \
--drop_threshold 0 \
--output_dir finetuned_bge-m3 \
--deepspeed examples/finetune/ds_stage0.json \
--overwrite_output_dir \
--learning_rate 5e-6 \
--num_train_epochs 2 \
--per_device_train_batch_size 2 \
--dataloader_drop_last True \
--warmup_ratio 0.1 \
--gradient_checkpointing \
--logging_steps 20 \
--save_steps 5000 \
--negatives_cross_device \
--temperature 0.05 \
--sentence_pooling_method cls \
--normalize_embeddings True \
--kd_loss_type m3_kd_loss \
--unified_finetuning True \
--use_self_distill True \
--fix_encoder False \
--self_distill_start_step 500
Bash
복사
In /FlagEmbedding/FlagEmbedding/finetune/embedder/encoder_only/m3/trainer.py file, I added functions within EncoderOnlyEmbedderM3Trainer class that tracks and logs the loss during training to monitor training progress. Every 100 steps, it saves this history to a log file in JSON format.
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
self.loss_history = []
# Create loss file in output directory
self.log_file = os.path.join(self.args.output_dir, "training_loss.json")
def _save(self, output_dir: Optional[str] = None, state_dict=None):
# rest of the function
self._save_loss_log()
def compute_loss(self, model, inputs, return_outputs=False, **kwargs):
loss, outputs = super().compute_loss(model, inputs, return_outputs=True);
self.loss_history.append({
"step": self.state.global_step,
"loss": loss.item(),
})
if self.state.global_step % 100 == 0:
self._save_loss_log()
return (loss, outputs) if return_outputs else loss
def _save_loss_log(self):
if self.is_world_process_zero():
os.makedirs(os.path.dirname(self.log_file), exist_ok=True)
with open(self.log_file, 'w') as f:
json.dump(self.loss_history, f, indent=2)
Python
복사
5. Model Evaluation
Within FlagEmbedding there are several evaluation datasets in place, including datasets like MIRACL and MLDR that supports Korean. However, because not all data is the same and training on one dataset doesn’t necessarily guarantee a performance increase for another, we may want to assess finetuned models on the test data of our training dataset. Thus, in this practice we will be using a custom dataset.
To use a custom dataset for evaluation, you need three files: corpus.jsonl, test_queries.jsonl, test_qrels.jsonl. Each file must be structured as below.
example corpus.jsonl
{"id": "101", "title": "", "text": "사업자등록증을 분실했을 경우, 관할 세무서 또는 홈택스를 통해 재발급 신청이 가능합니다."}
{"id": "102", "title": "", "text": "부가세 신고 중 잘못 입력한 경우, 신고 기한 내에 수정신고서를 제출하여 수정 가능합니다."}
{"id": "103", "title": "", "text": "퇴직금 계산 시 기본급, 상여금, 연차수당 등이 포함됩니다."}
{"id": "104", "title": "", "text": "외국인 근로자의 소득세 신고를 위해 여권, 비자, 급여 명세서가 필요합니다."}
{"id": "105", "title": "", "text": "정부 지원금 신청은 중소벤처기업부 사이트에서 신청서를 작성하고 관련 서류를 제출해야 합니다."}
{"id": "106", "title": "", "text": "대표자 변경 시 은행 계좌 변경을 위해 신분증, 사업자등록증, 변경 증명 서류가 필요합니다."}
JSON
복사
example test_queries.jsonl
{"id": "1", "text": "사업자등록증 분실 시 대처 방법은?"}
{"id": "2", "text": "부가세 신고 시 실수로 잘못 입력한 경우 수정할 수 있나요?"}
{"id": "3", "text": "퇴직금 계산 기준에 포함되는 항목은 무엇인가요?"}
{"id": "4", "text": "외국인 근로자 세금 신고 서류는 무엇이 필요한가요?"}
JSON
복사
example test_qrels.jsonl
{"qid": "1", "docid": "101", "relevance": 1}
{"qid": "2", "docid": "102", "relevance": 1}
{"qid": "3", "docid": "103", "relevance": 1}
{"qid": "4", "docid": "104", "relevance": 1}
JSON
복사
Below is an example script that will produce these three files based on the QA dataset the training dataset was created from.
import json
import hashlib
def get_document_id(text):
return "doc_" + hashlib.md5(text.encode('utf-8')).hexdigest()[:12]
def generate_files(data, output_path):
corpus_entries = []
query_entries = []
qrel_entries = []
seen_texts = set()
for idx, item in enumerate(data, start=1):
query_id = str(idx)
# Add query entry
query_entries.append({
"id": query_id,
"text": item["query"]
})
# Add positive document and relevance judgment
pos_text = item["pos"][0]
pos_doc_id = get_document_id(pos_text)
if pos_text not in seen_texts:
seen_texts.add(pos_text)
corpus_entries.append({
"id": pos_doc_id,
"title": "",
"text": pos_text
})
qrel_entries.append({
"qid": query_id,
"docid": pos_doc_id,
"relevance": 1
})
# Add negative documents and relevance judgments
for neg_text in item["neg"]:
neg_doc_id = get_document_id(neg_text)
if neg_text not in seen_texts:
seen_texts.add(neg_text)
corpus_entries.append({
"id": neg_doc_id,
"title": "",
"text": neg_text
})
qrel_entries.append({
"qid": query_id,
"docid": neg_doc_id,
"relevance": 0
})
# corpus.jsonl
with open(f"{output_path}/corpus.jsonl", "w", encoding="utf-8") as f:
for entry in corpus_entries:
f.write(json.dumps(entry, ensure_ascii=False) + "\n")
# test_queries.jsonl
with open(f"{output_path}/test_queries.jsonl", "w", encoding="utf-8") as f:
for entry in query_entries:
f.write(json.dumps(entry, ensure_ascii=False) + "\n")
# test_qrels.jsonl
with open(f"{output_path}/test_qrels.jsonl", "w", encoding="utf-8") as f:
for entry in qrel_entries:
f.write(json.dumps(entry, ensure_ascii=False) + "\n")
test_data = []
with open("toy_finetune_data_minedHN.jsonl", "r", encoding="utf-8") as f:
for line in f:
if line.strip():
data = json.loads(line.strip())
test_data.append(data)
output_path = "./output_path"
generate_files(test_data, output_path)
Python
복사
After generating the files, put them into a single directory and run the command below.
python -m FlagEmbedding.evaluation.custom \
--eval_name your_data_name \
--dataset_dir ./your_data_path \
--splits test \
--corpus_embd_save_dir ./your_data_name/corpus_embd \
--output_dir ./your_data_name/search_results \
--search_top_k 50 \
--cache_path ./cache/data \
--overwrite True \
--eval_output_method markdown \
--eval_output_path ./your_data_name/eval_results.md \
--eval_metrics ndcg_at_1 ndcg_at_3 ndcg_at_5 ndcg_at_10 recall_at_1 recall_at_3 recall_at_5 recall_at_10 \
--embedder_name_or_path finetuned_bge-m3 \
--embedder_model_class encoder-only-m3 \
--devices cuda:1 \
--cache_dir ./cache/model
Bash
복사
Notice that we use two main evaluation metrics, ndcg and recall.
•
NDCG evaluates both relevance and position in the top-k.
•
Recall measures whether the system retrieves all relevant document within the top-k
•
We don’t use precision in this case because precision measures the proportion of retrieved documents that are relevant. You may recall that our dataset was created from a QA set and have a single positive document for every query. With only one pos, precision becomes less informative with greater k.
6. Troubleshooting
Error while creating shared memory segment
•
On installation, if you are using Docker, this is an example configuration for the docker run command that bypasses the shared memory error.
docker run --shm-size=4g -it -d --gpus all --name flagembed -v $(pwd):/workspace <IMAGE_NAME>
Bash
복사
Bus error (core dumped)
•
Setting CUDA devices to cuda:1 instead of cuda:0 fixed the issue
•
Unsure of why this was the case